Uncertainty in deep learning
- Overview:
- Reminder of Bayesian inference
- Uncertainty at the output of DNN
- Uncertainty in latent layer
- Bayesian Neural Networks
- refs:
- https://www.youtube.com/watch?app=desktop&v=lhwk4ESlyMA&feature=youtu.be
- Goal1: compute how reliable the model output is
- we know the output of MLP classifier = \(p(y|x)\)
- but we know it overestimates the probability of its prediction
- Better method: conformal prediction sets
- paper
- use a held-out dataset to estimate proba that model is correct
- for any target confidence, returns a set of possible labels
Bayes vs. DNNs
- Basic DNNs do not consider the process that generates data:
- They consider only fixed-point data samples
- Given one point \(x\), they compute one output \(y\)
- Exemple:
- inputs = day, month, location
- output = temperature
- A fixed point is not enough: there’s a lot of variability
- wind, previous weather in neighbouring places…
- We’d prefer mean + stdev
- 1st source of uncertainty:
- many factors are unknown
- 2nd source of uncertainty:
- our model is limited
- ex: input = history + geo-grid of T°,P,W,rain,lum
- output still erronous
- Bayesian networks
- model the distribution of \(y\) given \(x\)
- Get uncertainty of the prediction
- Enable sampling various good outputs
- You may include prior knowledge
- e.g., T° increase over years because of global warming
- But BN are
- less powerful than DNNs
- very costly to train (in general)
- Can we do the same with DNNs ?
Brief reminder of Bayesian models
- Bayesian models assume all vars are generated from a process:
- Variable \(X\) is sampled from a distribution, which is usually assumed parametric: \[X \sim Mult(\theta)\]
- \(\theta\) usually trained with MLE, MAP or Bayesien inference
- Maximum Likelihood
- maximum likelihood <=> cross-entropy
- data follow a true distribution \(P^*(X)\), approximated by parametric distrib \(P(X;\theta)\)
- MLE: \(\theta^* = \arg\max_\theta P(X;\theta)\)
- Assume iid samples:
- \(\theta^* = \arg\max_\theta \prod_i P(X_i;\theta) = \arg\max_\theta \sum_i \log P(X_i;\theta)\)
- \(\theta^* = \arg\min_\theta E_{X \sim P^*(X)} \left[\log \frac 1 {P(X;\theta)} \right]\)
- which is the equation of the cross-entropy
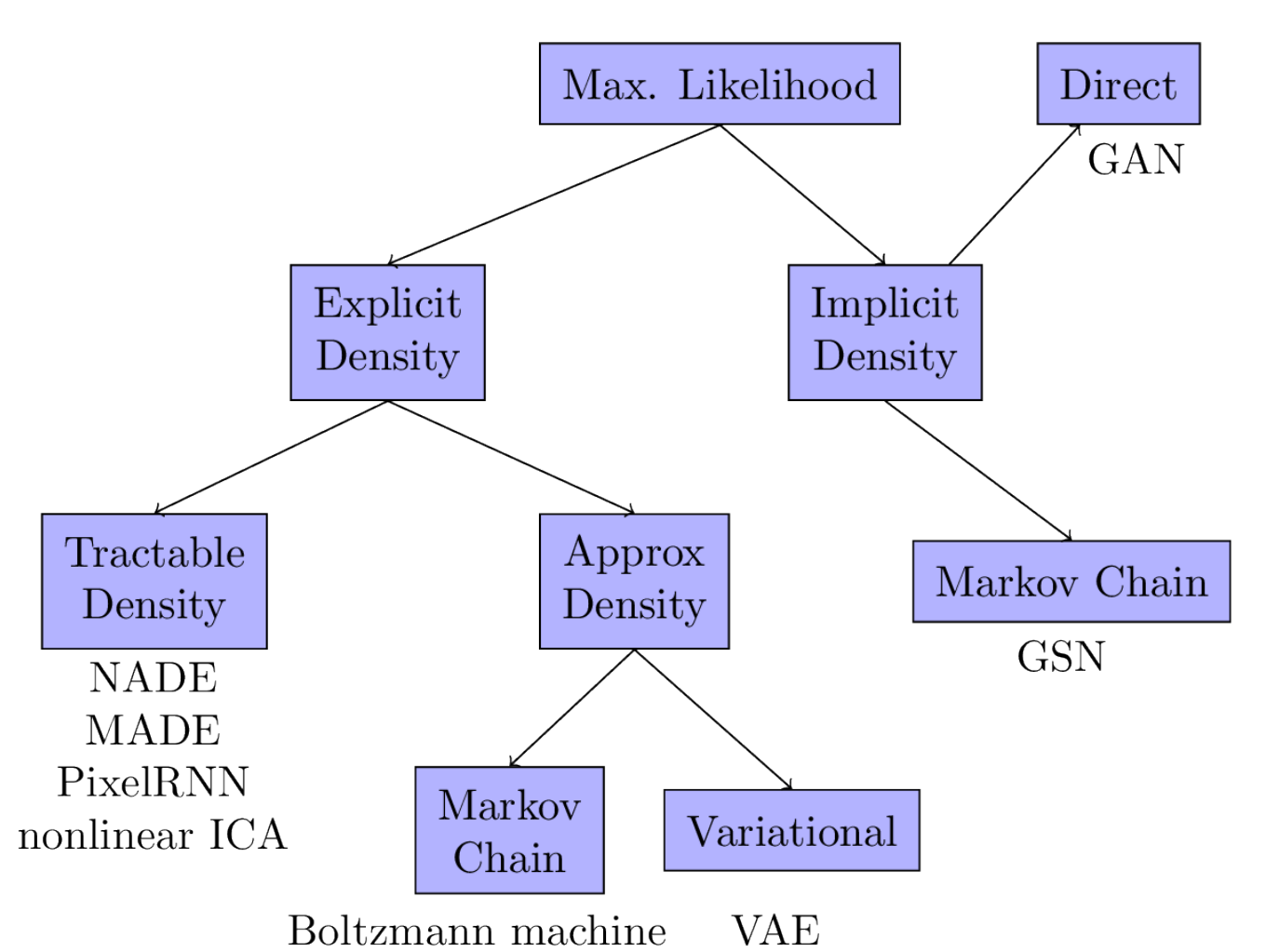
Limits of Maximum Likelihood
- Only relies on the evidence
- Does not take into account prior knowledge
- but why bother about priors?
- Ex: we know that, in July, T° is usually positive
- MLE will “rediscover” this fact through data
- bad use of evidence / training time
- much more efficient to tell it beforehand
- Prior knowledge makes better use of few data
- Bayesian models assume that we have some knowledge about the
distribution before observing anything
- prior distribution \(P(\theta)\)
- Now \(\theta\) is a random variable !
- If the prior knowledge is good enough, then we don’t need data at all
- Combining prior knowledge + data: (1)
- We want to find the best \(\theta\) according to both prior and likelihood
- maximum a posteriori: \(\max_\theta P(\theta|X) = \max_\theta P(X|\theta) P(\theta)\)
- L2 regularization <=> MAP with a Gaussian prior on the weights
- Combining prior knowledge + data: (2)
- Estimate the full posterior: observations (evidence) come in to
modify this prior belief:
- Transforms the prior \(P(\theta)\) into the posterior \(P(\theta|X)\)
- = Bayesian Inference
SGD == Approx. Bayesian Inference
https://arxiv.org/abs/1704.04289
- constant-rate SGD simulates a Markov chain with a stationary distrib
- possible to adjust the LR of constant SGD to best match the stationary distrib to a posterior
Distributions in DNNs
- If we use a vanilla DNN, do we have any information about the distribution of possible outputs?
- How can we modify the vanilla DNN to get more information?
- Vanilla DNN:
- Can we measure uncertainty of the output?
- DNN that outputs a distribution (Mixture Density Network)
- DNN with a latent Random Variable
- RBM, DBN
- VAE
- Bayesian DNN
Can we still get a distribution out of a vanilla DNN ?
- Famous examples: generative model:
- A generative model samples from a density of probability
- VAE: the latent \(z\) is randomly sampled from a Gaussian distribution
- GAN: the generator is fed with a random input
Softmax approximates posterior
- Multi-class classification:
- Objective = classify an input \(x\)
- e.g., does an utterance express joy, anger, sadness… ?
- Celebs pictures: who is she/he ?
- One output neuron per class
- each output neuron gives a score (logits)
- normalized by softmax –> probability
- … which probability ?
- Objective = classify an input \(x\)
- (Bishop,94) proves that a MLP trained with cross-entropy (and infinite data) approximates the posterior \(P(y|x)\)
- But in practice, not very reliable to estimate uncertainty
- Gal & Ghahramani (2016) show that a model can be uncertain in its predictions even with a high softmax output.
- Calibration
- Bishop proposed another analysis to estimate output distrib with MSE
Gaussian approximation
(Bishop94):
- assume infinite data \((x,y)\), a DNN \(f(\cdot;\theta)\); the MSE error is:
\[\int \int (f(x;\theta) - y)^2 p(x,y) dx dy\]
\[(f(x;\theta) - y)^2 = (f(x;\theta) - y + E[y|x] - E[y|x])^2\]
\[\begin{eqnarray} (f(x;\theta) - y)^2 &=& (f(x;\theta) - E[y|x])^2 + (E[y|x]-y)^2 + \\ && 2(f(x;\theta) - E[y|x])(E[y|x]-y) \end{eqnarray}\]
\[\begin{eqnarray} (f(x;\theta) - y)^2 &=& (f(x;\theta) - E[y|x])^2 + E[y|x]^2 + y^2 -2E[y|x]y +\\ && 2f(x;\theta)E[y|x] -2f(x;\theta)y - 2E[y|x]^2 + 2E[y|x]y \end{eqnarray}\]
\[\begin{eqnarray} (f(x;\theta) - y)^2 &=& (f(x;\theta) - E[y|x])^2 + y^2 + 2f(x;\theta)E[y|x] - \\ && 2f(x;\theta)y -E[y|x]^2 \end{eqnarray}\]
Consider the first term in \(2f(x;\theta)...\) into the integral:
\[\begin{eqnarray} \int 2f(x;\theta)E[y|x] p(y|x)p(x) dy &=& 2f(x;\theta) p(x) \int E[y|x] p(y|x) dy \\ &=& 2f(x;\theta) E[y|x] p(x) \end{eqnarray}\]
for the other term: \[\begin{eqnarray} \int 2f(x;\theta)y p(y|x)p(x)dy &=& 2f(x;\theta) p(x) \int y p(y|x)dy \\ &=& 2f(x;\theta) E[y|x] p(x) \end{eqnarray}\]
so \[\begin{eqnarray} \int (f(x;\theta) - y)^2 p(x,y)dy &=& p(x)(f(x;\theta) - E[y|x])^2 +\\ && p(x)(E[y^2|x] - E[y|x]^2) \end{eqnarray}\]
so the MSE error is \[\int (f(x;\theta) - E[y|x])^2 p(x)dx + \int (E[y^2|x] - E[y|x]^2)p(x)dx\]
The MSE error only depends on the model parameters through the first term, and it is minimum when \(f(x;\theta) = E[y|x]\)
- Consequently, if we want to model uncertainty of the DNN output with
a Gaussian, we can interpret that, after training, the DNN:
- outputs the expected value (mean)
- The variance of the error (=uncertainty) can be computed from the corpus (second term)
- OK, we have way to estimate the uncertainty from the corpus
- but can we let the DNN computes it itself ?
Gaussian mixture approximation
- (Bishop, 94) proposes the “Mixture Density Network”:
- the input X is passed into an MLP with 3 outputs:
- \(K\) mixing values \(\alpha\), which are further passed into a softmax
- \(K\) means \(\mu\)
- \(K\) variances \(\sigma\), which are further passed into some positive-only function
- the input X is passed into an MLP with 3 outputs:
- the final output of the model is a mixture of Gaussians: \(\sum_k \alpha_k N(x;\mu_k,\sigma_k)\)
- The output is a probability distribution that represents the possible values of \(y\) given \(x\)
- So if you want just one value \(y\), you have to sample
- See Bishop’s report on MDN
DNNs with latent random variable
- We have seen how to model the parameters of a GMM with a DNN
- It is possible to model parametric distributions inside a DNN, but only for a Gaussian.
- This may seem very limited, but the following DNN layers may transform this Gaussian into any other distribution!
VAE
- Explicitly model a distribution
- Encoder transforms input \(x\) into a distribution \(p(z)\sim N(\mu,\sigma)\)
- Decoder transforms a sample \(z\sim p(z)\) into the output \(\hat x\)
- We want to train the VAE with MLE, but the likelihood is hard to compute: \[\log p(x) = \log \int p(z)p(x|z)dz\]
- variational principle:
- convert difficult computation into optim. problem
- we chose a variational family \(q(z|x) \in Q\)
- How to get the ELBO (Evidence Lower BOund): \[\log p(x) = \log \int \frac
{q(z|x)}{q(z|x)}p(x,z)dz = \log E_q \frac {p(x,z)}{q(z|x)}\]
- the log of a sum is not easy to differentiate…
- Much better to get the sum of the log:
- Jensen’s inequality gives the ELBO: \[\geq E_{q(z|x)} \log \frac {p(x,z)}{q(z|x)}\]
Note: when we maximize the lower bound, we actually make \(q\) closer to \(p\): \[\max_q E_{q(z|x)} \log \frac {p(x,z)}{q(z|x)} = \log p(x) - \min_q D(q(z|x)||p(z|x))\]
So MLE involves now a double optimization (\(\hat p\) is the empirical corpus): \[\max_{p,q} E_{\hat p(x)}E_{q(z|x)} \log \frac {p(x,z)}{q(z|x)}\]
Let us consider encoder parameters \(\phi\) and decoder parameters \(\theta\) \[\max_{\theta,\phi} E_{\hat p(x)}E_{q_\phi(z|x)} \log \frac {p(z)p_\theta(x|z)}{q_\phi(z|x)}\]
SGD involves computing \[\nabla_\phi E_{q_\phi(z|x)} \log \frac {p(z)p_\theta(x|z)}{q_\phi(z|x)}\]
- hard: cannot be expressed as an expected value of a gradient
- let’s make it not dependent on \(\phi\): reparameterization trick
Instead of sampling from \(q\), we sample from a fixed distribution \(p(\epsilon)\): \[\nabla_\phi E_{q_\phi(z|x)} \log \frac {p(z)p_\theta(x|z)}{q_\phi(z|x)} = \nabla_\phi E_{p(\epsilon)} \log \frac {p(T(\epsilon))p_\theta(x|T(\epsilon))}{q_\phi(T(\epsilon)|x)}\]
which is possible when \(q_\phi(z|x)\) is a conditional diagonal Gaussian model with: \[p(\epsilon)=N(0,1)\] \[T(\epsilon)=\mu_\phi(x) + \epsilon \cdot \sigma_\phi(x)\]
- We now have: \[\nabla_\phi E_{p(\epsilon)} \log \frac {p(T(\epsilon))p_\theta(x|T(\epsilon))}{q_\phi(T(\epsilon)|x)}= E_{p(\epsilon)} \nabla_\phi \log \frac {p(T(\epsilon))p_\theta(x|T(\epsilon))}{q_\phi(T(\epsilon)|x)}\]
- can be estimated with Monte Carlo!
In practice
- given \(x\), the encoder computes \(\mu_\phi(x)\) and \(\sigma_\phi(x)\)
- sample an \(\epsilon \sim N(0,1)\)
- build the latent representation \(z=\mu_\phi(x) + \epsilon \cdot \sigma_\phi(x)\)
- pass \(z\) to the decoder \(\hat x = p_\theta(x|z)\)
- optimize sum of:
- standard loss (MSE, cross-ent) for \(\theta\): \(l(\hat x, x)\)
- regularizer for \(\phi\): \(KL(q_\phi(z|x)||N(0,1))\)
- and for a given \(x\): \[KL(q_\phi(z|x)||N(0,1))= \mathbb{KL}\left( \mathcal{N}(\mu, \sigma) \parallel \mathcal{N}(0, 1) \right)\] \[ = \sigma^2 + \mu^2 - \log \sigma - \frac{1}{2}\]
General pb: non-diff
- Doubly stochastic system:
- stochastic component: sampling in the DNN
- stochastic optimization: sampling a batch in SGD
- Let’s define \(\hat x \sim p_\theta(x)\) the (random) output of the DNN
- \(f\) is the loss fct, we can estimate the expected loss with MC: \[E_{p_\theta}[f(x)] \simeq \frac 1 N \sum_i^N f(\hat x^{(i)})\]
- for SGD, we need the gradient of the expected loss: \[\nabla_\theta E_{p_\theta}[f(x)] = \nabla_\theta \int p_\theta(x) f(x)dx = \int \nabla_\theta p_\theta(x) f(x)dx\]
- but cannot be cast as an expectation, because \(\nabla_\theta p_\theta(x)\) is not a density (may be <0)
- same thing for the ELBO: \[\nabla_\theta ELBO(q) = \nabla_\theta E_q [\log p(x,z) - \log q(z)]\]
- 2 solutions:
- score function estimator (REINFORCE)
- pathwise gradient estimator (Reparameterization)
- Sol 1: score function estimator
- a.k.a. REINFORCE, or likelihood ratio estimator
- unbiased, no restriction on \(p_\theta(x)\) and \(f(x)\)
- but it has large variance: might give very different \(\theta\)
- def: score = \(\nabla_\theta \log p_\theta(x)\)
\[\nabla_\theta E_{p_\theta}[f(x)] = \int \nabla_\theta p_\theta(x) f(x)dx\] \[=\int p_\theta(x) \frac {\nabla_\theta p_\theta(x)}{p_\theta(x)} f(x)dx\] \[=\int p_\theta(x) \nabla_\theta \log p_\theta(x) f(x)dx\] \[=E_{p_\theta}[\nabla_\theta \log p_\theta(x) f(x)]\]
Can be estimated with Monte Carlo !
- For the ELBO: \[\nabla_\theta E_q [\log p(x,z) - \log q(z)] = \nabla_\theta E_q [f_\theta(z)]\] \[ = \int \nabla_\theta (q_\theta(z) f_\theta(z))dz\] \[ = \int \nabla_\theta q_\theta(z) f_\theta(z)dz + q_\theta(z) \nabla_\theta f_\theta(z)dz\] \[ = E_q [\nabla_\theta \log q_\theta(z)(\log p(x,z)-\log q_\theta(z))]\]
Can be estimated with Monte Carlo ! But pb: large variance
- Sol 2: pathwise gradient estimator
- instead of direct sampling \(\hat x \sim p_\theta(x)\)
- indirect sampling: \(\hat x = t_\theta(\hat\epsilon)\) with \(\hat\epsilon \sim p(\epsilon)\)
- \(p(\epsilon)\) = base distribution
- \(t_\theta(\hat\epsilon)\) = sampling path (deterministic)
- example: \(x=t_\theta(\epsilon)=\mu + \sigma\epsilon\) and \(N(\epsilon;0,1)\)
- law of unconscious statistician: \[E_{p_\theta(x)}[f(x)] = E_{p(\epsilon)}[f(t_\theta(\epsilon))]\]
\[\nabla_\theta E_{p_\theta(x)} [f(x)] = \nabla_\theta E_{p(\epsilon)}[f(t_\theta(\epsilon))]\] \[\nabla_\theta E_{p_\theta(x)} [f(x)] = \int p(\epsilon)\nabla_\theta f(t_\theta(\epsilon))d\epsilon\] \[\nabla_\theta E_{p_\theta(x)} [f(x)] = \int p(\epsilon)\nabla_x f(x) \nabla_\theta t_\theta(\epsilon)d\epsilon\]
- The loss must be differentiable!
- pathwise estimator has better variance, but it requires
- differentiable loss
- rewriting the sampling of \(x\) with a deterministic function with all parameters
Bayesian DNN
So far:
- Standard DNN= trained with MLE (point estimate)
- L2-Regularized DNN= trained with MAP (Gaussian prior, point estimate)
- What would be a better training procedure ?
- posterior inference ! But very difficult.
- old approx 1 = Laplace’s method
- old approx 2 = MCMC (long convergence)
- recent approx = BNN with variational inference
- BNN = weights are Random Variable (stochastic networks)
- tuto: cf https://arxiv.org/pdf/2007.06823.pdf
- toolkit = pyro
- we want the full predictive distribution
\[p(y|D,x) = \int p(y|w,x) p(w|D) dw\]
- several ways to approximate it (with different names)
\[p(y|D,x) = \int p(y|w,x) p(w|D) dw\]
- approximate predictive distribution with MCMC:
- sample a few possible w
- combine the resulting model
- … == deep ensembles !
- Monte Carlo Dropout
- proposed by Gal & Ghahramani (2016)
- dropout = averaging ensemble of many different networks
- apply dropout also during testing
- == approx Bayesian inference
- Stochastic Weight Averaging
- combine weights of the same network at different stages of training
- SWA-Gaussian further approx the posterior distrib locally using only info from SGD
- refs: https://medium.com/neuralspace/bayesian-neural-network-series-post-1-need-for-bayesian-networks-e209e66b70b2
- https://www.cc.gatech.edu/~hic/8803-Fall-09/slides/8803-09-lec07.pdf for Laplace approximation that leads to Bayesian DNN
Practice: VAE
- Goal = train and analyze a VAE on MNIST
- first train an auto-encoder on MNIST: run this code to train the AE
import torch; torch.manual_seed(0)
import torch.nn as nn
import torch.nn.functional as F
import torch.utils
import torch.distributions
import torchvision
import numpy as np
import matplotlib.pyplot as plt; plt.rcParams['figure.dpi'] = 200
device = "cpu"
class Encoder(nn.Module):
def __init__(self, latent_dims):
super(Encoder, self).__init__()
self.linear1 = nn.Linear(784, 512)
self.linear2 = nn.Linear(512, latent_dims)
def forward(self, x):
x = torch.flatten(x, start_dim=1)
x = F.relu(self.linear1(x))
return self.linear2(x)
class Decoder(nn.Module):
def __init__(self, latent_dims):
super(Decoder, self).__init__()
self.linear1 = nn.Linear(latent_dims, 512)
self.linear2 = nn.Linear(512, 784)
def forward(self, z):
z = F.relu(self.linear1(z))
z = torch.sigmoid(self.linear2(z))
return z.reshape((-1, 1, 28, 28))
class Autoencoder(nn.Module):
def __init__(self, latent_dims):
super(Autoencoder, self).__init__()
self.encoder = Encoder(latent_dims)
self.decoder = Decoder(latent_dims)
def forward(self, x):
z = self.encoder(x)
return self.decoder(z)
def train(autoencoder, data, epochs=20):
opt = torch.optim.Adam(autoencoder.parameters())
for epoch in range(epochs):
for x, y in data:
x = x.to(device) # GPU
opt.zero_grad()
x_hat = autoencoder(x)
loss = ((x - x_hat)**2).sum()
print("train",epoch,loss.item())
loss.backward()
opt.step()
if epoch%1==0:
plot_latent(autoencoder, data)
# plot_reconstructed(autoencoder)
return autoencoder
latent_dims = 2
autoencoder = Autoencoder(latent_dims).to(device) # GPU
data = torch.utils.data.DataLoader(
torchvision.datasets.MNIST('./data',
transform=torchvision.transforms.ToTensor(),
download=True),
batch_size=128,
shuffle=True)
autoencoder = train(autoencoder, data)- Then plot the latent space with:
def plot_latent(autoencoder, data, num_batches=100):
for i, (x, y) in enumerate(data):
z = autoencoder.encoder(x.to(device))
z = z.to('cpu').detach().numpy()
plt.scatter(z[:, 0], z[:, 1], c=y, cmap='tab10')
if i > num_batches:
plt.colorbar()
break
plt.show()- Then plot the “reconstructed” space with:
def plot_reconstructed(autoencoder, r0=(-5, 10), r1=(-10, 5), n=12):
w = 28
img = np.zeros((n*w, n*w))
for i, y in enumerate(np.linspace(*r1, n)):
for j, x in enumerate(np.linspace(*r0, n)):
z = torch.Tensor([[x, y]]).to(device)
x_hat = autoencoder.decoder(z)
x_hat = x_hat.reshape(28, 28).to('cpu').detach().numpy()
img[(n-1-i)*w:(n-1-i+1)*w, j*w:(j+1)*w] = x_hat
plt.imshow(img, extent=[*r0, *r1])- look at the areas not seen in the latent space
- Then modify the AE into a VAE with the class at next slide
- TODO = compute the KL term of the loss
- then add this KL term into the loss in the train() function:
loss = ((x - x_hat)**2).sum() + autoencoder.encoder.kl- VAE class:
class VariationalEncoder(nn.Module):
def __init__(self, latent_dims):
super(VariationalEncoder, self).__init__()
self.linear1 = nn.Linear(784, 512)
self.linear2 = nn.Linear(512, latent_dims)
self.linear3 = nn.Linear(512, latent_dims)
self.N = torch.distributions.Normal(0, 1)
self.N.loc = self.N.loc.cuda() # hack to get sampling on the GPU
self.N.scale = self.N.scale.cuda()
self.kl = 0
def forward(self, x):
x = torch.flatten(x, start_dim=1)
x = F.relu(self.linear1(x))
mu = self.linear2(x)
sigma = torch.exp(self.linear3(x))
z = mu + sigma*self.N.sample(mu.shape)
self.kl = # TODO: compute and store the KL loss here
return z- compare the size and shape of the latent spaces of AE and VAE
- compare the reconstructions in areas not covered by AE
Practice: Pyro
- Install:
pip install pyro-ppl- Doc: https://readthedocs.org/projects/pyro-ppl/downloads/pdf/dev/
- slides: https://sshkhr.github.io/files/ProbProgtutorial.pdf
Sampling from a distribution:
import pyro.distributions as dist
from pyro import sample
from torch import tensor
coinflip = sample("coinflip", dist.Bernoulli(probs=0.5))
noisy_sample = sample("noisy_sample", dist.Normal(loc=0, scale=1))- creates named random variables
Computing log-likelihood:
print(dist.Normal(0, 1).log_prob(tensor(0.35)).exp()) # 0.3752Implementing a generative model: \(P(sleeptime,tired) = P(sleeptime|tired)P(tired)\)
def genmod():
amTired = sample("being_tired", dist.Bernouilli(0.7))
if amTired: sleepTime = sample("sleep_time", dist.Normal(8,1))
else: sleepTime = sample("sleep_time", dist.Normal(6,1))
return sleepTime- How can you answer questions ?
- joint proba of a sample: \(P(sleeptime=2,tired=1)\)
- joint distrib: \(P(sleeptime,tired)\)
- marginal proba of a sample: \(P(tired=1)\)
- marginal distrib: \(P(tired)\)
- Useful method 1: trace
- collect all (var name, sampled value)
- Useful method 2: condition
- Set the value of a variable instead of sampling
- Get the joint probability of sample (as a pytorch computation graph !):
from pyro.poutine import trace
from pprint import pprint
# trace() internally calls genmod()
tr = trace(genmod).get_trace()
pprint({
name: {
'value': props['value'],
'prob': props['fn'].log_prob(props['value']).exp()
}
for (name, props) in tr.nodes.items()
if props['type'] == 'sample'
})
print(tr.log_prob_sum().exp())- Get the marginal distribution over each variable:
import pandas as pd
import matplotlib.pyplot as plt
traces = []
for _ in range(1000):
tr = trace(sleep_model).get_trace()
values = {
name: props['value'].item() for (name, props) in tr.nodes.items() if props['type'] == 'sample'
}
traces.append(values)
pd.DataFrame(traces).hist()
plt.show()from pyro import condition
cond_model = condition(genmod, {
"being_tired": tensor(1.),
"sleep_time": tensor(10.)
})
trace(cond_model).get_trace().log_prob_sum().exp()- Conditional proba: what is the probability that I’m tired given that I slept 6.5 hours ?
- Can be done with Bayes rule: \[P(tired=1|sleep=6.5) = \frac {P(t=1,s=6.5)}{\sum_t P(t,s=6.5)}\]
- Pb1: with many variables, exponential growth of terms in the sum
- Pb2: with continuous variables, integral instead of sum
- approximate inference (cf. CS 228 of Stanford)
- MCMC or variational
- pyro focuses on VI
- Toy model: Gaussian with 1 parameter (= pytorch parameter tensor) initialized at 0.3:
from pyro import param
mu = param("mu",tensor(0.3))
mod = dist.Normal(mu,1)MLE training
- Toy data: we observe 1 value \(x=5\)
- Compute prob = likelihood for this value
- SGD update to maximize prob:
prob = mod.log_prob(5)
prob.backward()
mu.data += mu.grad- Rewrite with our previous methods:
def model():
mu = param("mu", tensor(0.))
return sample("x", dist.Normal(mu, 1))
cond_model = condition(model, {"x": 5})
tr = trace(cond_model).get_trace()
tr.log_prob_sum().backward()
mu = param("mu")
mu.data += mu.grad- Note that each variable is uniquely identified by its name !
- With pytorch optimizer:
from torch.optim import Adam
def model():
mu = param("mu", tensor(0.))
return sample("x", dist.Normal(mu, 1))
model() # Instantiate the mu parameter
cond_model = condition(model, {"x": tensor(5.)})
optimizer = Adam([param("mu")], lr=0.01)
mus, losses = [], []
for _ in range(100):
tr = trace(cond_model).get_trace()
# minimize negative log probability
prob = -tr.log_prob_sum()
prob.backward()
optimizer.step()
optimizer.zero_grad()
losses.append(prob.item())
mus.append(param("mu").item())
pd.DataFrame({"mu": mus, "loss": losses}).plot(subplots=True)
plt.show()Posterior Inference
- so far, we’ve done MLE
- we need posterior inference to answer: what is the probability that I’m tired given that I slept 6.5 hours ?
- we cannot simply set “sleeptime=6.5” and sample values of “tired”, because the generative model first sample “tired”, and then set “sleeptime”.
- we would need to sample both variables, and filter sleeptime=6.5 afterwards: intractable !
- approx. inference with VI
- var. distrib \(q\) is called “guide” in pyro:
- we may approx. our target posterior \(p(tired|sleeptime=6.5)\) with a dirac at 1:
def sleep_guide():
sample("being_tired", dist.Delta(1.))- we can compute ELBO (KL-div btw variational and posterior dist.) given a guide: \[E_{t\sim q} [\log P(t,s=6.5) - \log P_q(t)]\]
def elbo(guide, cond_model):
dist = 0.
# the expected value is a sum over possible values of "tired"
for t in [0., 1.]:
# we define the fct to compute the proba of a given t from a given model
log_prob = lambda f: trace(condition(f, {"being_tired": tensor(t)})).get_trace().log_prob_sum()
# the marginal from the guide
guide_prob = log_prob(guide)
# the joint from the model
cond_model_prob = log_prob(cond_model)
term = guide_prob.exp() * (cond_model_prob - guide_prob)
if not torch.isnan(term): dist += term
return dist
underslept = condition(genmod, {"sleep_time": 6.5})
elbo(sleep_guide, underslept)- we can now make the ELBO smaller:
pyro.clear_param_store()
def sleep_guide():
# Constraints ensure facts always remain true during optimization,
# e.g. that the parameter of a Bernoulli is always between 0 and 1
valid_prob = constraints.interval(0., 1.)
t = param('qt', tensor(0.8), constraint=valid_prob)
sample('being_tired', dist.Bernoulli(t))
# Run once to register the param calls with Pyro
sleep_guide()
adam = Adam([param('qt').unconstrained()], lr=0.005)
param_vals = []
for _ in range(2000):
# We can use our elbo function from earlier and compute its gradient
loss = -elbo(sleep_guide, underslept)
loss.backward()
adam.step()
adam.zero_grad()
param_vals.append({k: param(k).item() for k in ['qt']})
pd.DataFrame(param_vals).plot(subplots=True)- pb: if we use a continuous variable, ELBO requires to compute an integral: intractable
- we can approximate this integral by sampling several values, or just one !
from pyro.poutine import replay
def elbo_approx(guide, cond_model):
# sample one value from the guide to approximate the sum
guide_trace = trace(guide).get_trace()
model_trace = trace(replay(cond_model, guide_trace)).get_trace()
return model_trace.log_prob_sum() - guide_trace.log_prob_sum()- replay() == condition() but with a trace
- tip: when VI does not work, check the gradient !
- when gradient unstable, you may use surrogate objective:
def elbo_better_approx(guide, cond_model):
guide_trace = trace(guide).get_trace()
model_trace = trace(replay(cond_model, guide_trace)).get_trace()
elbo = model_trace.log_prob_sum() - guide_trace.log_prob_sum()
# "detach" means "don't compute gradients through this expression"
return guide_trace.log_prob_sum() * elbo.detach() + elbo- A simpler version of all this:
from pyro.optim import Adam
from pyro.infer import SVI, Trace_ELBO
adam = Adam({"lr": 0.005})
svi = SVI(underslept, sleep_guide, adam, loss=Trace_ELBO())
param_vals = []
for _ in range(2000):
svi.step()
param_vals.append({k: param(k).item() for k in ["qt"]})- Exercices:
- proba of being tired given that I slept 6.5 hours ?
- compare with “slept 9.5 hours” ? does it follow intuition ?
- Ex. to manipulate the methods:
- compute ELBO with other guides and check the values are ranked according to intuition
- plot the gradient of being_tired during training
- Another example: do the tuto with ‘fair coins’ here
- designing guides:
- flexible enough to approx. the distrib well
- with continuous variables for gradient descent
ref: https://willcrichton.net/notes/probabilistic-programming-under-the-hood/
Pyro and DNN
- Pyro HiddenLayer implements a linear NN layer + non-linear
activation
- with Normal prior in the weights
- with local reparameterization trick to speed up inference
- instead of sampling all NxN weights (W), we sample the N outputs of each layer (W.X)
MNIST in pyro: https://github.com/paraschopra/bayesian-neural-network-mnist